Description
This competition is the ideal starting point for you to learn the in’s and out’s of how federated machine learning code competitions on the tracebloc platform work. Think of this competition as a fun space to try out all the features we offer.
The key distinction between this code competition and others found on tracebloc is that it doesn't involve sensitive information. However, we encourage you to handle the data as if it were sensitive, using only the resources provided us. This will help you become accustomed to the exciting competitions that await you in the future.
The goal of this competition is to take some work from the shoulders of doctors and classify medical x-ray mammography images using your machine learning models. If you want to get to know the dataset, simply have a look at the "Data" and "Explorative Data Analysis" tabs.
Once you’re ready to join the fun, click on the "join competition" button.
The Challenge
Did you know that breast cancer is the most common cancer found in women worldwide?
Sadly, breast cancer prevention options are limited, which makes early detection incredibly important to increase the chances of a successful treatment and cure. Mammography is currently the most effective method for detecting breast cancer in women aged 50 to 69 years. Mammography is a medical imaging test utilizing low energy X-rays. During the test, the breast is compressed between two plates and an X-ray image is taken.
The critical point in this challenge is that every X-ray image contains a suspicious mass found by a radiologist. Your challenge will be to use machine learning techniques to accurately separate the malignant masses from the benign masses.
The Data
Our competition features the CBIS-DDSM dataset (Curated Breast Imaging Subset of DDSM), an updated and standardized version of the Digital Database for Screening Mammography (DDSM).
The DDSM is a comprehensive database of 2,620 scanned film mammography studies that contains benign and malignant cases with verified pathology information. This dataset is widely regarded as a valuable tool in the development and testing of decision support systems for breast cancer detection.
CBIS-DDSM (Curated Breast Imaging Subset of DDSM), includes decompressed images, data selection and curation by trained mammographers and pathologic diagnosis for training data, formatted like modern computer vision data sets.
Join us in this exciting code competition and help to find new ways of saving lives by further improving breast-cancer screening!
Resources
- Sawyer-Lee, R., Gimenez, F., Hoogi, A., & Rubin, D. (2016). Curated Breast Imaging Subset of Digital Database for Screening Mammography (CBIS-DDSM) (Version 1) [Data set]. The Cancer Imaging Archive. https://doi.org/10.7937/K9/TCIA.2016.7O02S9CY
Evaluation
Here you’ll find the evaluation metric your model's performance will be measured with, as well as a short example and explanation. The main objective of the competition is to improve that score by as much as possible.
For this competition, you are tasked with binary classification and need to improve your models F1 score on the test dataset.
The F1 score is the harmonic mean between precision and recall and can be computed like this (for an example ground truth and model prediction) (click here for reference)
Code Implementation
# Example Ground Truth and Model Prediction
ground_truth = [1, 1, 1, 0, 0, 1, 0, 1, 0]
prediction = [1, 1, 0, 0, 1, 1, 0, 1, 0]
# implementation using sklearn
from sklearn import metrics
metrics.f1_score(ground_truth, prediction)
# basic implementation
def true_positive(ground_truth, prediction):
tp = 0
for gt, pred in zip(ground_truth, prediction):
if gt == 1 and pred == 1:
tp +=1
return tp
def true_negative(ground_truth, prediction):
tn = 0
for gt, pred in zip(ground_truth, prediction):
if gt == 0 and pred == 0:
tn +=1
return tn
def false_positive(ground_truth, prediction):
fp = 0
for gt, pred in zip(ground_truth, prediction):
if gt == 0 and pred == 1:
fp +=1
return fp
def false_negative(ground_truth, prediction):
fn = 0
for gt, pred in zip(ground_truth, prediction):
if gt == 1 and pred == 0:
fn +=1
return fn
def recall(ground_truth, prediction):
tp = true_positive(ground_truth, prediction)
fn = false_negative(ground_truth, prediction)
prec = tp/ (tp + fn)
return prec
def precision(ground_truth, prediction):
tp = true_positive(ground_truth, prediction)
fp = false_positive(ground_truth, prediction)
prec = tp/ (tp + fp)
return prec
def f1(ground_truth, prediction):
p = precision(ground_truth, prediction)
r = recall(ground_truth, prediction)
f1_score = 2 * p * r/ (p + r)
return f1_score
f1(ground_truth, prediction)
Timeline
This competition will run all year and around the clock, but that won’t usually be the case with other competitions.
- Announced: The announcement date, is the point in time where the dataset or competition information are shared with the data science community.
- Start: The specific day and time when participants are able to start working on the data science competition.
- Team Merger Deadline: The last date by which participants can decide to join forces and work together as a team in the competition.
- Final submission deadline: The date by when the final 2 submissions need to be submitted. These two final submission from each team will be used to create the final leaderboard
- Final Results: The rankings or scores of all participants or teams, determined after all submissions are evaluated according to the competition's scoring system.
All deadlines are at 11:59 PM UTC on the corresponding day unless otherwise noted. The competition organizers reserve the right to update the contest timeline if they deem it necessary.
Acknowledgements
- Lee, R. S., Gimenez, F., Hoogi, A., Miyake, K. K., Gorovoy, M., & Rubin, D. L. (2017). A curated mammography data set for use in computer-aided detection and diagnosis research. In Scientific Data (Vol. 4, Issue 1). Springer Science and Business Media LLC. https://doi.org/10.1038/sdata.2017.177
-
- Clark, K., Vendt, B., Smith, K., Freymann, J., Kirby, J., Koppel, P., Moore, S., Phillips, S., Maffitt, D., Pringle, M., Tarbox, L., & Prior, F. (2013). The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository. In Journal of Digital Imaging (Vol. 26, Issue 6, pp. 1045–1057). Springer Science and Business Media LLC. https://doi.org/10.1007/s10278-013-9622-7
Data Details
Generally,
here you’ll find general information about the competition dataset. The data itself is kept federated within our partners and tracebloc’s infrastructure to ensure the privacy of the data. As this is competition is for introductory purposes, the privacy of the data doesn’t really need to be protected. However, this won’t be the case for most of our other competitions.
If you want to learn more about how federated learning works, have a look at this.
The Dataset
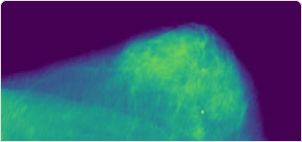
This excerpt from the CBIS-DDSM dataset contains 16000 images, with a train / test split of 0.75 / 0.25. The test dataset is further divided into a validation part (3000 images) and a test part (1000 images) for the final evaluation of your model. The images you can see here are 10 sample images that are representative but not part of the actual dataset you will be training your models on.
Your task is binary classification of the X-ray mammography images into the classes “benign” and “malignant”. The dataset contains images of both the right and left breast.
Evaluation
Evaluation
Here you’ll find the evaluation metric your model's performance will be measured with, as well as a short example and explanation. The main objective of the competition is to improve that score by as much as possible.
For this competition, you are tasked with binary classification and need to improve your models F1 score on the test dataset.
The F1 score is the harmonic mean between precision and recall and can be computed like this (for an example ground truth and model prediction) (click here for reference)
Code Implementation
# Example Ground Truth and Model Prediction
ground_truth = [1, 1, 1, 0, 0, 1, 0, 1, 0]
prediction = [1, 1, 0, 0, 1, 1, 0, 1, 0]
# implementation using sklearn
from sklearn import metrics
metrics.f1_score(ground_truth, prediction)
# basic implementation
def true_positive(ground_truth, prediction):
tp = 0
for gt, pred in zip(ground_truth, prediction):
if gt == 1 and pred == 1:
tp +=1
return tp
def true_negative(ground_truth, prediction):
tn = 0
for gt, pred in zip(ground_truth, prediction):
if gt == 0 and pred == 0:
tn +=1
return tn
def false_positive(ground_truth, prediction):
fp = 0
for gt, pred in zip(ground_truth, prediction):
if gt == 0 and pred == 1:
fp +=1
return fp
def false_negative(ground_truth, prediction):
fn = 0
for gt, pred in zip(ground_truth, prediction):
if gt == 1 and pred == 0:
fn +=1
return fn
def recall(ground_truth, prediction):
tp = true_positive(ground_truth, prediction)
fn = false_negative(ground_truth, prediction)
prec = tp/ (tp + fn)
return prec
def precision(ground_truth, prediction):
tp = true_positive(ground_truth, prediction)
fp = false_positive(ground_truth, prediction)
prec = tp/ (tp + fp)
return prec
def f1(ground_truth, prediction):
p = precision(ground_truth, prediction)
r = recall(ground_truth, prediction)
f1_score = 2 * p * r/ (p + r)
return f1_score
f1(ground_truth, prediction)
Timeline
General Info
This competition will run all year and around the clock, but that won’t usually be the case with other competitions.
- Announced: The announcement date, is the point in time where the dataset or competition information are shared with the data science community.
- Start: The specific day and time when participants are able to start working on the data science competition.
- Team Merger Deadline: The last date by which participants can decide to join forces and work together as a team in the competition.
- Final submission deadline: The date by when the final 2 submissions need to be submitted. These two final submission from each team will be used to create the final leaderboard
- Final Results: The rankings or scores of all participants or teams, determined after all submissions are evaluated according to the competition's scoring system.
All deadlines are at 11:59 PM UTC on the corresponding day unless otherwise noted. The competition organizers reserve the right to update the contest timeline if they deem it necessary.